Complex evolutionary roots of skin-barrier function in humans
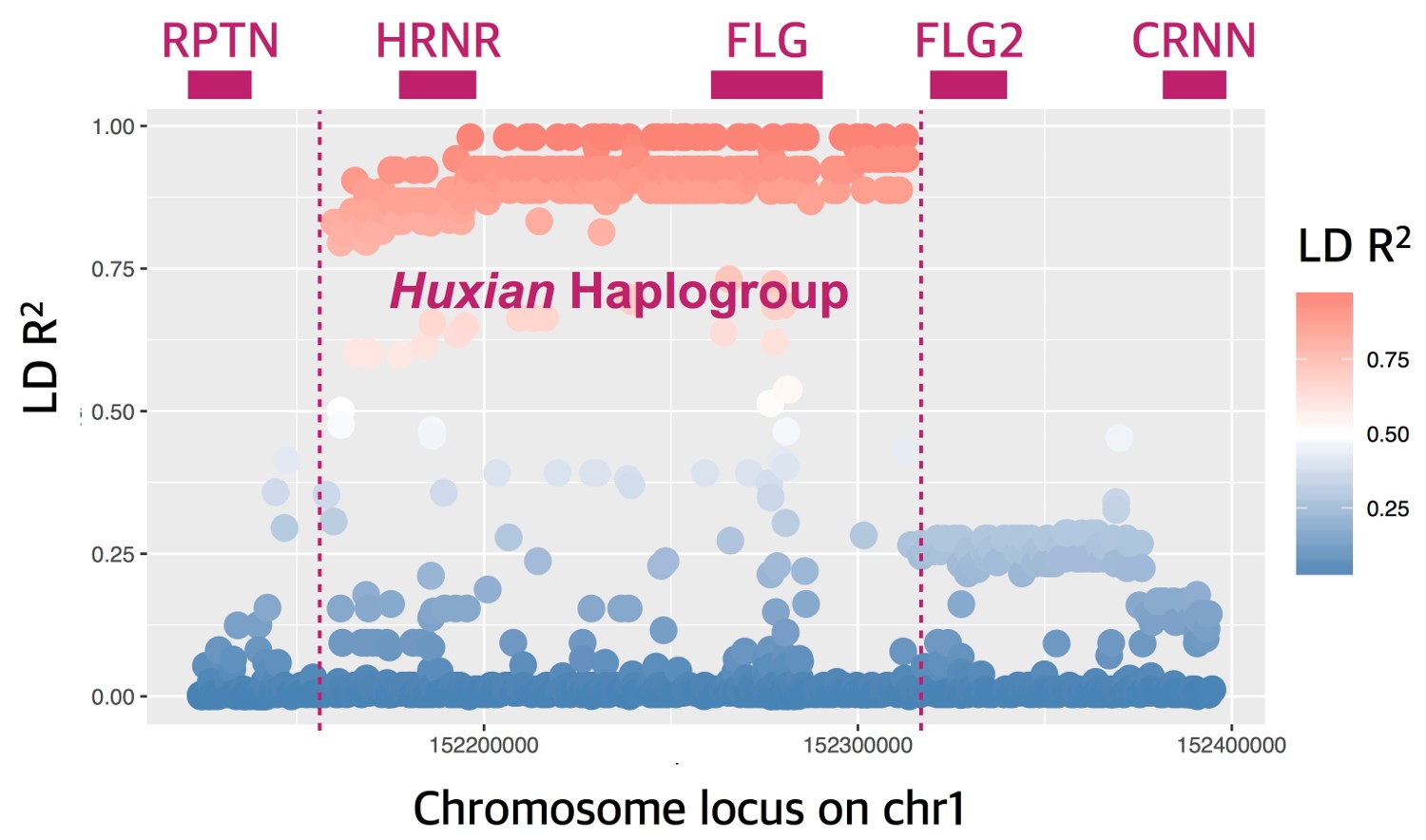
Rapid evolution of human skin has left evolutionary signatures in the genome. The filaggrin (FLG) is a well-studied gene essential for natural skin-barrier function in humans. The extensive genetic variation in this gene, especially common loss-of-function (LoF) mutations, has been strongly associated with atopic dermatitis susceptibility in multiple human populations. To investigate the evolution of this gene, we analyzed 2,504 human genomes and genotyped the copy number variation of filaggrin repeats within FLG in 126 individuals from diverse ancestral backgrounds. We were unable to replicate a recent study claiming that LoF of FLG is adaptive in northern latitudes with lower ultraviolet light exposure. Further, we integrated results from evolutionary and population genetic analyses, RNA expression data from multiple tissues, as well as molecular mechanisms of FLG function to reach multiple striking conclusions: (i) FLG LoF mutations are unusually common when compared to other genes in the genome, (ii) the most parsimonious explanation for this observation is that FLG LoF variants have no obvious fitness effects despite predisposing to skin disease, (iii) some of the atopic dermatitis susceptibility variants “hitchhike” a selective sweep involving neighboring hornerin (HRNR) gene, and (iv) the haplotype block that is under selection carry multiple functional variants, including those that change the expression level of multiple transcripts and microbiome diversity on the skin. Our results present multiple lines of evidence that involve complex evolutionary trajectories in shaping the skin-barrier function in humans.
Genomic Structural Variants in Anthropological Genetics

I studied genomic structural variants (SVs) affecting phenotypic variations among humans within an anthropological context. This work has provided a significant insight into incorporation of SVs for anthropological genetics studies. I demonstrated the anthropologically contextualized analysis of SVs for understanding the genetic bases of human phenotypic variation. In the line of postulating a methodological workflow, I examined 1000 Genomes deletion variants and identified 16 exonic deletions that are specific to the African continent. I analyzed two of these deletion variants (KAP-Del1 and KAP-Del2) affecting the keratin-associated protein (KAP) gene cluster in a locus-specific manner. This extensive KAP gene family has been associated with the evolution of hair in mammals and said to evolve under rapid, diversifying selection among mammals as a response to ever-changing ecological pressures regarding hair formation. This dynamic evolutionary history made the deletion variants affecting the KAP gene cluster, including KAP-Del1 and KAP-Del2, viable candidates to understand phenotypic variation in hair patterns among modern humans. My analysis revealed that these deletions may indeed affect phenotype and likely evolved under geography-specific positive selection.
Genetic diversity of ethno-religious populations of India
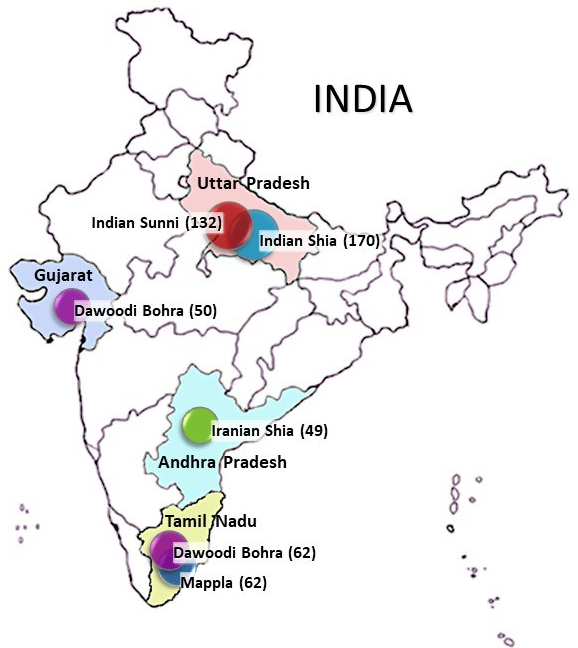
To delineate genetic ancestry and diversity of Indian Muslims, I examined several non-recombining genetics markers such as mtDNA and Y chromosome along with lactase persistence autosomal variants. My results support a model according to which the spread of Islam in India was predominantly cultural conversion associated with minor but still detectable levels of gene flow from outside, primarily from Iran and Central Asia, rather than directly from the Arabian Peninsula. My investigation on the variation in lactase persistence DNA variants corroborated the gene flow from Iran in the light of adaptation to the cultural practice of dairying. Further, I found traces of sub-Saharan African maternal lineage in a non-tribal population for the first time. Above all, this study revealed that geography and indigenous heritage manifests more profoundly in contrast to gene replacement processes attributable to the intrusion of peoples with a distinctive ethnicity. In addition, I also contributed to a number of human genetic diversity studies, in several ethnic population groups of India and Bangladesh.