I have been scrutinizing genome-wide genotypes and exome sequences of Kuwaiti population to understand their genetic diversity patterns and determine the population-specific genetic variants associated with diabetes and related diseases. In addition, I use evolutionary genetics approach to study natural selection and local adaptation to the desert environment.
Evolutionary roots of metabolic traits in an Arabian Peninsula population
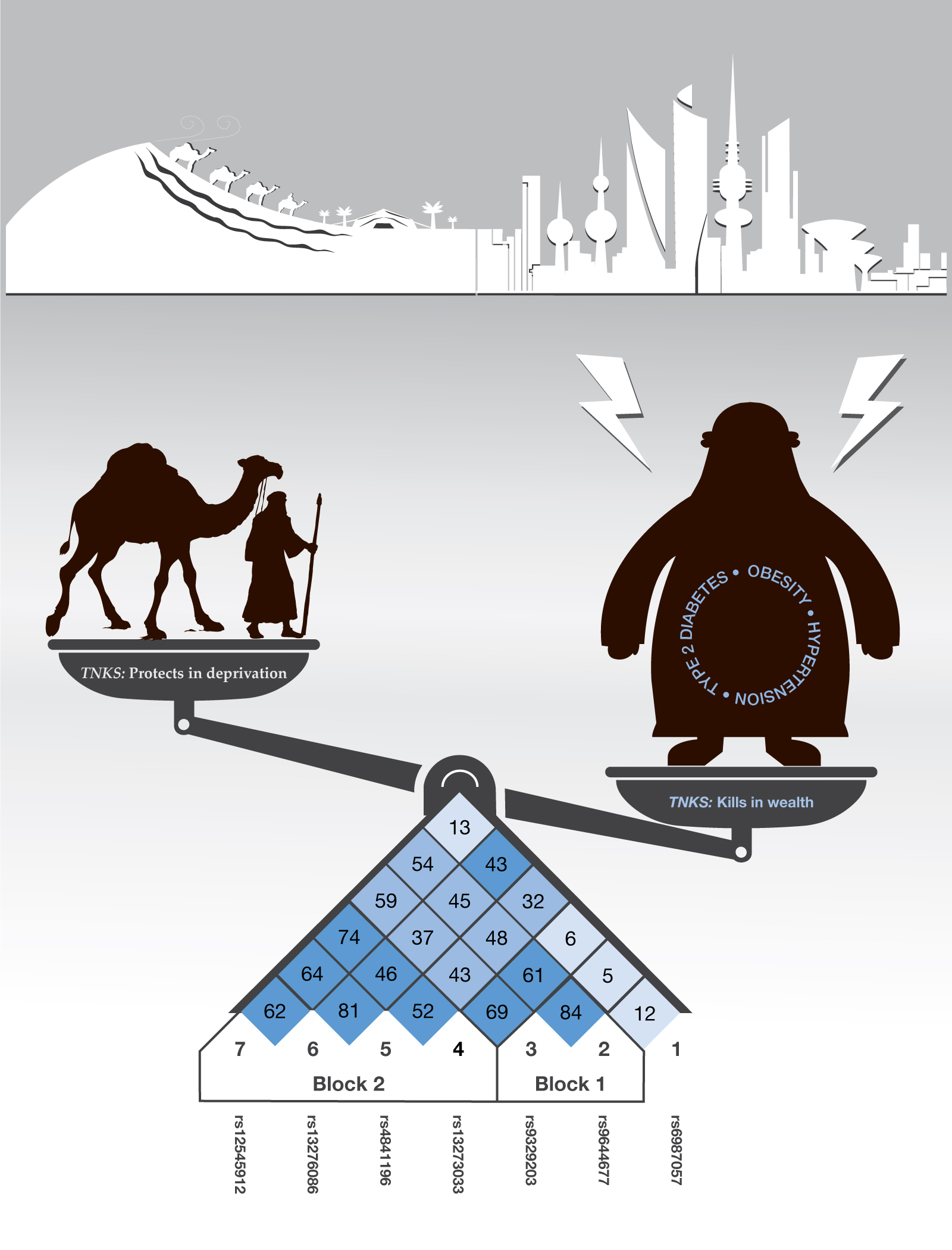
Following the out-of-Africa diaspora, the Arabian Peninsula has experienced several waves of human migrations despite the prevailing extreme and varying environmental conditions. The potential adaptation that shaped the extant human populations of the hot and dry environment of the Arabian Peninsula have been scarcely studied. We explored natural selection in the Arabian Peninsula region by analyzing 662,750 single nucleotide variants in 583 Kuwaiti and 96 Saudi Arabian individuals conducting multiple statistical tests, including integrated Haplotype Score (iHS), Cross Population Extended Haplotype Homozygosity (XP-EHH), Population Branch Statistics (PBS), and log-likelihood ratio scores (LLRS). Our integrative approach identified several genomic regions with strong signals of positive selection in both the populations, many of which contain loci associated with metabolic traits, asthma and blood-related phenotypes. We highlighted a haplotype overlapping TNKS region in chromosome 8, associated with metabolic traits and hypertension. The TNKS haplotype exemplifies a general trend in which a more rapid metabolism rate and hypertension have been selected in the Kuwaiti population, and potentially conferred some degree of fitness advantage to the Kuwaiti ancestors for surviving in the extremely dry and hot ecological environments, while posing a considerable health risk to present-day Kuwait populations.
Genetic diversity and affinities of the Kuwait population in the Greater Middle East region
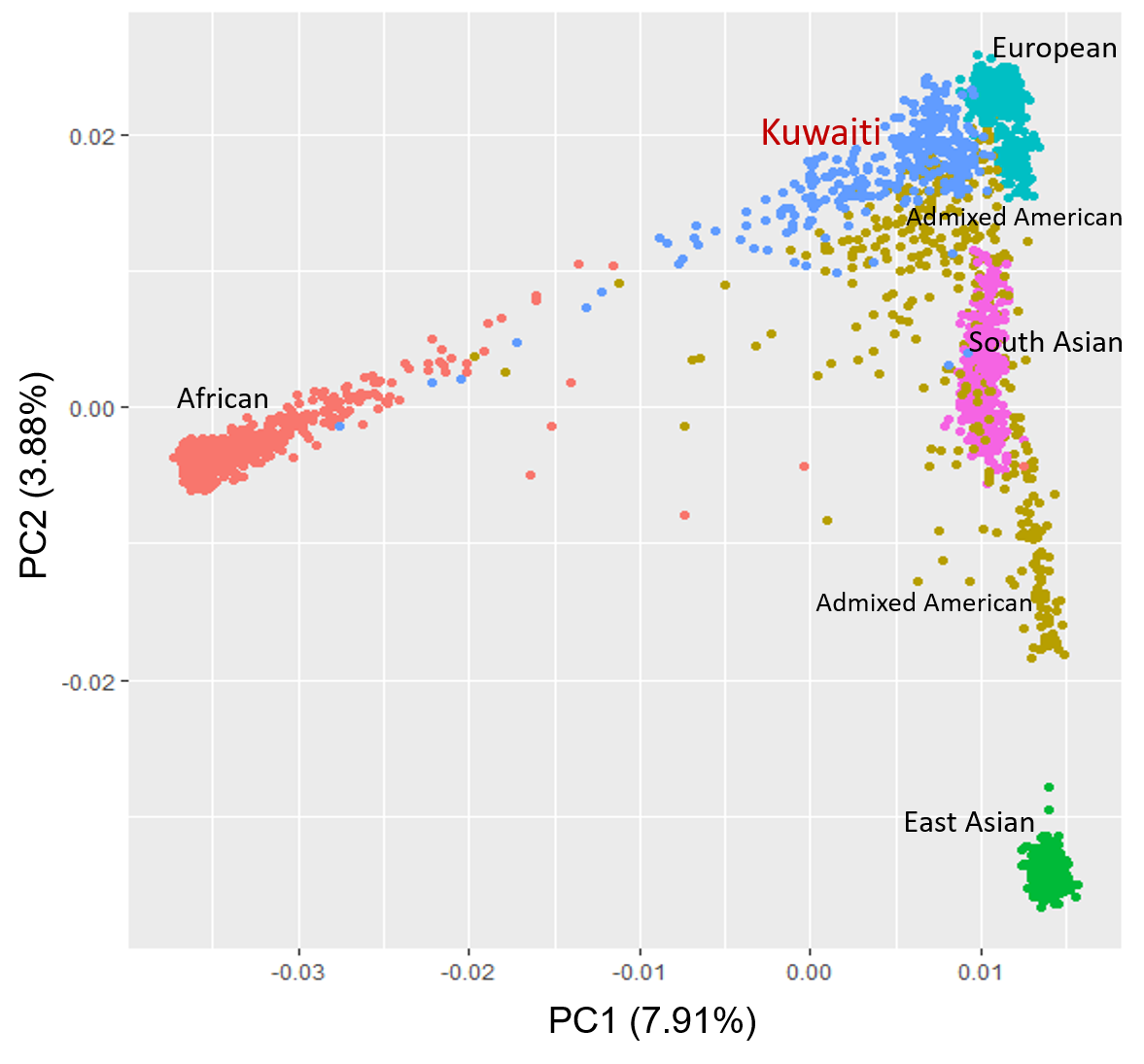
We conducted fine-scale population genetic analysis by analyzing about 620 genome-wide genotypes (244,688 SNPs) and 300 exome sequences of Kuwaiti individuals. To understand the genetic diversity patterns in the regional and global perspective, we merged the in-house dataset with pooled dataset from several published sources and 1000 Genomes project phase3 dataset containing about 1,564 individuals from 84 different continental populations and 2,504 individuals from 26 global populations, respectively. The admixture analysis detected the ancestral genetic elements from North Africa, Saudi Arabia and Caucasus in each of the three Kuwaiti subpopulation groups. Population stratification analysis presented the placement of Kuwaiti populations among the global populations. The comprehensive estimations for genetic differentiation, runs of homozygosity, identity by descent and haplotype-sharing concur showing similar genetic affinity and diversity of Kuwaiti populations in the greater Middle East region.
Energy balance and mitochondrial DNA variation in Kuwaiti populations
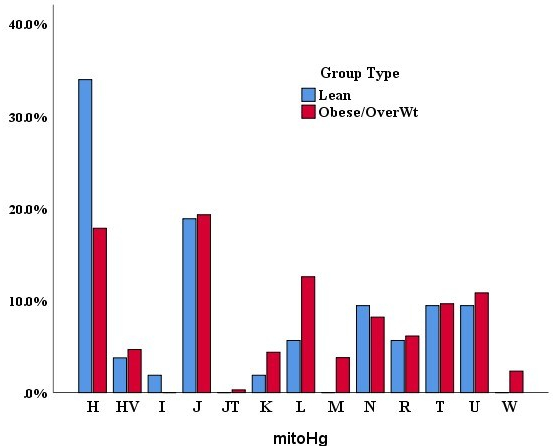
Kuwait represents three times higher (49.1%) the world average obesity rate (13%). Accordingly, Kuwaiti people are at high risk of health burdens like diabetes, cardiovascular disease and cancer. This condition is attributed to physically inactive lifestyle that leads to energy imbalance in the body, apparently linking to the functioning of the powerhouse of the cell, mitochondria. Considering the importance of mitochondrial DNA (mtDNA) variation in energy balancing process, we are analysing mtDNA genetic variation in two stages, (1) non-coding control region spanning about 1,200 base pairs; and (2) complete mitochondrial genome of size 16,569 base pairs. In the initial analyses, I found mtDNA variants likely to occur mostly in obese Kuwaiti individuals when compared to normal individuals. Further, haplotype and haplogroup -based analyses also indicated obesity specific scenario. The complete mitochondrial genome analysis is underway.